Expression of Proteins from Lactose-Inducible Vectors in E. coli
Overview
Teaching: 60 min
Exercises: 450 minQuestions
How can bacteria be used to express a foreign protein?
Objectives
Students will be able to distinguish between an expression vector before and after insertion of a protein’s coding sequence.
Students will be able to describe the following features of an expression vector: multiple cloning site, T7 promoter region, antibiotic resistance gene, purification tag(s), origin of replication.
Students will explain why a chosen expression vector allows for expression of the desired protein.
Students will understand the following steps required for bacterial expression of a protein of interest: transformation, antibiotic selection, bacterial growth, induction.
Students will use knowledge of the lac operon to explain how its use in recombinant protein expression.
Students will understand the modifications made to standard bacteriological growth medium to induce protein expression.
Students will describe advantages and limitations of a bacterial expression system as a source of protein relative to original tissue/cell origin.
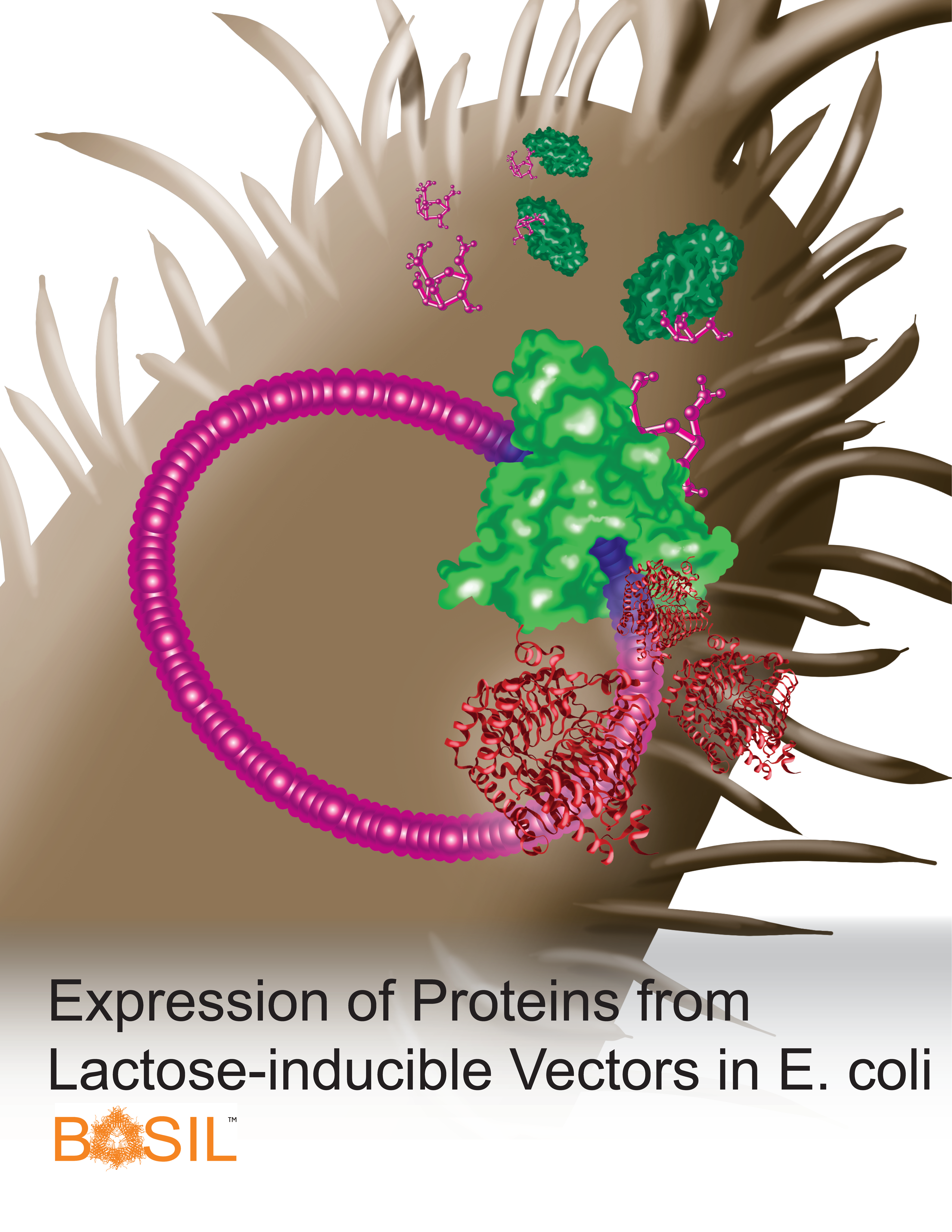 This module addresses how to use a bacterial expression system to produce a protein of interest for use in later experimental studies. The BASIL project uses structures of enzymes to predict their function. To test this predicted function, you must produce and purify the enzyme in the lab and then undertake a series of experiments. Enzymes may be isolated from the organism that naturally produces them, or they may be made by introducing a gene encoding the protein into a bacterial expression system. Bacterial expression systems are easy to work with and can produce a large amount of the desired protein in a short amount of time. By the end of this module, students will obtain bacterial cells containing their protein of interest that can be purified and then analyzed by a variety of experiments contained in other BASIL modules.
This module addresses how to use a bacterial expression system to produce a protein of interest for use in later experimental studies. The BASIL project uses structures of enzymes to predict their function. To test this predicted function, you must produce and purify the enzyme in the lab and then undertake a series of experiments. Enzymes may be isolated from the organism that naturally produces them, or they may be made by introducing a gene encoding the protein into a bacterial expression system. Bacterial expression systems are easy to work with and can produce a large amount of the desired protein in a short amount of time. By the end of this module, students will obtain bacterial cells containing their protein of interest that can be purified and then analyzed by a variety of experiments contained in other BASIL modules.
Module Resources
Key Points
Introduction of a foreign gene into E. coli allows researchers to express proteins of interest.
Protein expression can be controlled using knowledge of operators and repressors in E. coli.